

Novel Mediators of Amyloid Precursor Protein Signaling This section is compiled by Frank M. Painter, D.C.
Send all comments or additions to: Frankp@chiro.org
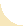


FROM: Journal of Neuroscience 2009 (Dec 16); 29 (50): 15703–15712 ~ FULL TEXT
Andrzej Swistowski, Qiang Zhang, Mark E. Orcholski, Danielle Crippen, Cathy Vitelli, Alexei Kurakin and Dale E. Bredesen
Buck Institute for Age Research,
Novato, California 94945, USA.Multiple recent reports implicate amyloid precursor protein (APP) signaling in the pathogenesis of Alzheimer's disease, but the APP-dependent signaling network involved has not been defined. Here, we report a novel consensus sequence for interaction with the PDZ-1 and PDZ-2 domains of the APP-interacting proteins Mint1, Mint2, and Mint3 (X11alpha, X11beta, and X11gamma), and multiple novel interactors for these proteins, with the finding that transcriptional coactivators are highly represented among these interactors. Furthermore, we show that Mint3 interaction with a set of the transcriptional coactivators leads to nuclear localization and transactivation, whereas interaction of the same set with Mint1 or Mint2 prevents nuclear localization and transactivation. These results define new mediators of the signal transduction network mediated by APP.
From the Full Text Article:
Introduction
Alzheimer's disease (AD) affects more than five million Americans, yet it is currently without any truly effective treatment. Although interest has in the past focused primarily on amyloid-β (Aβ) contained in plaques, it has become clear that plaque formation is not required for the Alzheimer's phenotype, at least in transgenic mouse models of AD [Hsia et al., 1999]. A number of previous reports have demonstrated toxicity of Aβ based on chemical and physical effects on the cell, such as lysosomotropic detergent-like effects [Glabe, 2001], metal binding [Barnham and Bush, 2008], and generation of reactive oxygen species [Butterfield, 2003]. Recent, complementary results argue that signaling events, some of which are mediated by APP itself, also play a crucial role in the development of the AD phenotype [Galvan et al., 2006; Zhao et al., 2006; Townsend et al., 2007; Ma et al., 2008; Nguyen et al., 2008]. Furthermore, APP has recently been shown to be a receptor for netrin-1 [Lourenço et al., 2009], and to give rise to N-APP, a ligand for DR6 (death receptor 6) [Nikolaev et al., 2009]. Therefore, it is of interest to determine the signaling networks centered around APP and to determine which signaling pathways and proteins are crucial for the development of the Alzheimer's phenotype.
The Mint1/X11 family has three members: Mint1, Mint2, and Mint3, also known as X11, X11L, and X11L2 or, alternatively, as X11α, X11β, and X11γ, respectively. All three members display a centrally located PTB (phosphotyrosine binding) domain, through which they bind to the intracellular domain of APP. They also contain two contiguous C-terminal postsynaptic density-95/Discs large/zonula occludens-1 (PDZ) domains [Rogelj et al., 2006]. Mint1/X11 family members have been implicated in synaptic vesicle exocytosis [Butz et al., 1998], protein transport and trafficking [Setou et al., 2000], neuronal plasticity [Ashley et al., 2005], and signaling [Tomita et al., 2000; Biederer et al., 2002]. Importantly, both in vitro and in vivo studies have indicated that the Mint1/X11 family members are involved in the regulation of APP processing and Aβ production [Mueller et al., 2000; Lee et al., 2003, 2004; King et al., 2004; King and Scott Turner, 2004; Ho et al., 2008; Saito et al., 2008]. Therefore, the identification of novel binding partners of the Mint1/X11 family members may lead to the elucidation of detailed mechanisms of AD pathology and provide novel potential targets for therapeutic development.
Target-assisted iterative screening (TAIS) is a phage display-based approach that allows for the rapid identification of multiple interactors—including high-affinity, intermediate-affinity, and low-affinity interactors—for a given protein interaction domain [Kurakin and Bredesen, 2002]. Here, we report the results of a study in which we used TAIS and identified 46 novel interactors of the PDZ-1 and PDZ-2 domains of Mint1. We further confirmed that two of the interactors, TAZ and YAP, associate with APP through Mint1/X11 family proteins. We then found that APP, Mint3, and TAZ/YAP form transcriptionally active triple protein complexes, and that the regulation of APP processing modulates the activities of Mint3/TAZ and Mint3/YAP complexes. These studies suggest that TAZ and YAP may serve as downstream mediators of APP signaling.
Materials and Methods
DNA constructs and phage display library.
The 16-mer random peptide library was generated using the T7 phage display library construction kit from Novagen. The glutathione S-transferase (GST) fusion protein expression constructs of the Mint1 PDZ-1 (amino acids 648-746) and PDZ-2 (740-825) were generated by PCR cloning into pGEX2TK expression vector (GE Healthcare). To avoid potential protein aggregation problems after bacterial expression, Cys 654 and Cys 742 (flanking both ends of PDZ1 and the N terminus of PDZ2) were replaced with Ser residues. The constructs were verified by sequencing. Expression and protein purification of GST fusions were performed according to the manufacturer's instructions. DNA constructs pMst(Gal4), pMst-APP (APP-Gal4), pMst-APP* (APP*-Gal4), pG5E1B-Luc (Gal4 reporter plasmid), pCMV-LacZ (β-galactosidase control plasmid), pCMV-Mint1, pCMV-Mint2, and pCMV-Mint3 were kindly provided by Dr. Patrick Mehlen (Université de Lyon, Lyon, France) and Dr. Thomas Südhof (University of Texas Southwestern Medical Center, Dallas, TX). Constructs pcDNA4-His-MaxB-hYAP1 and pEGFP-C3-hYAP1 were kindly provided by Dr. Marius Sudol (Weis Center for Research, Danville, PA). Construct pEF-N-FLAG-TAZ was kindly provided by Dr. Michael Yaffe (Massachusetts Institute of Technology, Boston, MA) and Dr. Iain Farrance (University of Maryland School of Medicine, Baltimore, MD). Construct pBIND-Gal4-DBD-X11L2 (Mint3) was kindly provided by Dr. Toshiharu Suzuki (Hokkaido University, Sapporo, Japan). Construct pcDNA3-APP695 was described previously [Lu et al., 2000].
Antibodies.
Mouse anti-Mint1 monoclonal antibody was obtained from BD Biosciences. Rabbit anti-Mint3 polyclonal antibody was obtained from Abcam. M2-anti-Flag mouse monoclonal antibody and M2 antibody-conjugated agarose beads were obtained from Sigma-Aldrich. CT15 anti-APP antibody was a kind gift from Dr. Edward Koo (University of California, San Diego, La Jolla, CA).
Target-assisted iterative screening.
A detailed description of the target-assisted iterative screening (TAIS) method has been described previously [Kurakin et al., 2004]. Briefly, 30 µg of a GST-PDZ domain fusion immobilized on Sepharose beads was blocked with 0.5% bovine serum albumin (BSA) in TBS-T (Tris-buffered saline, pH 7.4, plus 0.1% Tween 20) and incubated with a phage-displayed peptide library aliquot (~108–109 pfu). After 120 min of incubation at room temperature (RT), the beads were thoroughly washed with TBS-T and bound phages were eluted with 200 µl of 1% SDS for 15 min at RT. After elution, the phages were immediately mixed with a molten 0.6% top agarose containing host cells and plated onto two prewarmed 150 mm agar plates. When phage plaques became visible, the plates were cooled down for 30 min at 4°C and overlaid with 132 mm nitrocellulose membranes (Whatman Schleicher and Schuell) for 5 min. After plaque lift, the membranes were blocked in 1% BSA in TBS for 1 h at RT and incubated overnight in 25 ml of TBS-T on a rocker at 4°C with 10 µg of the target PDZ domain that had been cleaved from the GST moiety, biotinylated, and complexed with STRAP (streptavidin–alkaline phosphatase) at a ratio of 4:1. After extensive washing with TBS-T, positive plaques were developed on the membranes with insoluble AP (alkaline phosphatase) substrate BCIP (5-bromo-4-chloro-3-indolyl phosphate)/NBT (nitroblue tetrazolium) (Sigma-Aldrich). Individual positive plaques were identified on the plates, and phages from these plaques were propagated separately in the appropriate host for production of individual phage lysates. The identities of isolated phage-displayed peptides were inferred by sequencing the library-specific DNA inserts amplified by PCR from the T7 phage display vector.
Phage ELISA.
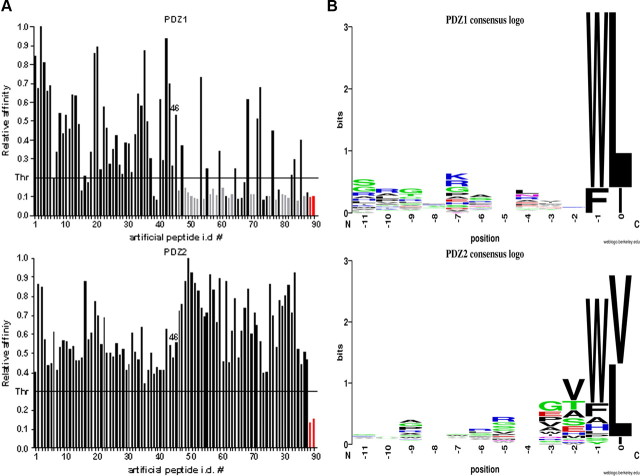
Figure 1A GST-PDZ domain fusion proteins were immobilized on microtiter ELISA plates (Corning Life Sciences) at 1 µg per well in 200 µl of 0.1 m NaHCO3, pH 8.0, overnight at 4°C. After protein coating, plates were blocked by adding 150 µl of 1% BSA in TBS for 1 h at RT and washed five (×1 ml) times with TBS-T. One hundred microliters of freshly prepared individual phage lysate was added to the ELISA plate wells and incubated for 1 h at RT. Unbound phages were washed away with TBS-T, and the number of retained phages was determined with monoclonal T7 phage-specific antibodies (Novagen; 1:1000) followed by monoclonal anti-mouse antibodies conjugated to horseradish peroxidase (HRP) (GE Healthcare; 1:1000). The individual phage ELISA kinetics was followed and quantified colorimetrically using soluble HRP substrate (ABTS/H2O2). ELISA readings were taken on a SpectraMAX190 plate reader (Molecular Devices) at 405 nm. To ensure reproducibility, each individual phage ELISA was repeated three times in three separate experiments. The representative binding histograms are shown in Figure 1A.
Peptide ELISA.
The synthetic N-terminally biotinylated peptides corresponding to natural ligand sequences were purchased from JPT Peptide Technologies. Wells of microtiter plates were coated with 1 µg of GST-PDZ domain fusions, washed with TBS-T, and blocked with 1% BSA as described above for phage ELISA. Individual biotinylated peptides (80 ng) were preincubated in separate microtiter plates with 1 µg per well of streptavidin-HRP conjugate (Pierce) in 250 ml of TBS-T for 30 min at RT.
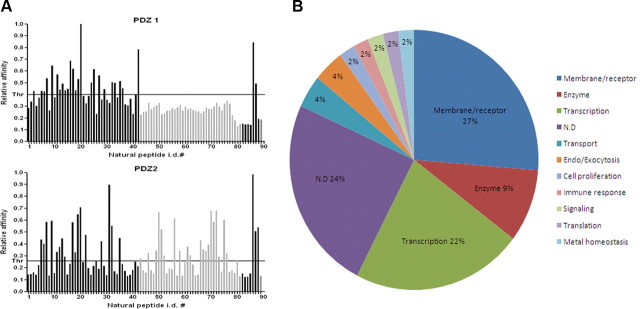
Figure 3A Fifty microliters of the peptide–streptavidin–HRP conjugate were added to 150 µl of TBS-T left in each coated well after the final wash of the protein immobilization/blocking procedure. Microtiter plates were incubated for 1 h at RT, and then washed five (×1 ml) times with TBS-T. The peptides retained were quantified colorimetrically by adding soluble HRP substrate (ABTS/H2O2) and measuring ELISA kinetic slopes. ELISA readings were taken on a SpectraMAX190 plate reader (Molecular Devices) at 405 nm. To ensure reproducibility, all peptide ELISA experiments were repeated at least three times in at least three separate experiments. The representative binding histograms are shown in Figure 3A.
Cell culture and coimmunoprecipitations.
HEK293T cells were cultured under standard conditions. Plasmid constructs were transiently transfected into HEK293T cells with Lipofectamine 2000 (Invitrogen). Coimmunoprecipitation and Western analysis were performed as previously described [Lu et al., 2000]. Briefly, 48 h after transfection, cells were harvested and lysed in NP-40 Cell Lysis Buffer (50 mm HEPES, pH 7.6, 125 mm NaCl, 5 mm EDTA, and 1% NP-40), and then, after centrifugation, incubated overnight with M2-anti-FLAG antibody-conjugated agarose beads (Sigma-Aldrich). The beads were subjected to five rounds of washing consisting of centrifugation, withdrawal of supernatant, and addition of fresh NP-40 Cell Lysis Buffer. During the final washing step, beads were resuspended in 1× LDS loading buffer (Invitrogen) with 50 mm DTT, and boiled at 100°C for 10 min. After SDS-PAGE and electrotransfer, Western blotting was performed using anti-Mint1, anti-Mint3, or anti-APP(CT15) antibodies. Thirty minutes of TBS-Tween wash were followed by incubation with secondary goat-anti-mouse (for Mint1 blotting) or goat-anti-rabbit (for Mint3 and APP blotting) antibodies. Another 30 min TBS-Tween wash was then followed by chemiluminescent detection (GE Healthcare).
Immunocytochemistry.
HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were trypsinized and plated into chamber slides at ~10% confluency. Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde for 20 min, and then permeabilized with 0.2% Triton X-100 for 15 min. For single staining, cells were blocked in PBS/10% donkey serum for 1 h at room temperature, incubated with primary antibody at 4°C overnight, and then stained with secondary antibody for 1 h at room temperature. For double staining, sequential staining protocol was applied. Cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) to visualize the nuclei. Images were captured on a laser-scanning confocal microscope (40×; Zeiss LSM510).
Luciferase transactivation assay.
HEK293T cells were cotransfected with five or six plasmids: (1) pG5E1B-luc, 0.3 µg; (2) pCMV-LacZ, 0.1 µg; (3) pMst-APP (APP-Gal4), pMst-APP* (APP*-Gal4), pBIND-Gal4-DBD-X11L2 (Mint3), 0.3 µg; (4) pCMV-Mint1, pCMV-Mint2, pCMV-Mint3, pcDNA3-APP695, 1.0 µg; (5) pEF-N-FLAG-TAZ, pcDNA4-His-MaxB-hYAP1, 1.0 µg. Where indicated, a sixth plasmid was cotransfected: pCMV-Mint1, pCMV-Mint2, 1.0 µg. For negative controls, the expression vector pcDNA3 was used without insert. Cells were harvested 48 h after transfection in 0.2 ml per well Cell Culture Lysis Buffer (Promega), and their luciferase and β-galactosidase activities were determined with the Promega luciferase assay kit and the Promega β-galactosidase assay kit, respectively. The luciferase activity was standardized by the β-galactosidase activity to control for transfection efficiency and general effects on transcription and further normalized for the transactivation observed in cells expressing APP-Gal4 or Mint3-Gal4 alone. Values shown are averages of transactivation assays performed in duplicate or triplicate and repeated at least three times for each cell type and constructs. All constructs were assayed in two or three cell lines, and representative results from one cell line are shown. Transfections were performed at 80–90% confluency in six-well plates using Lipofectamine 2000 (Invitrogen).
Results
Binding specificity profiles for PDZ domains of Mint1
To identify binding ligands of both PDZ domains of Mint1, we performed TAIS of a phage display peptide library, using GST-PDZ fusions as targets. We isolated and sequenced binding clones for each domain and obtained 46 and 42 unique sequences (referred to here as artificial peptides) for PDZ-1 and PDZ-2, respectively. Sequences were aligned and analyzed for their homology (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Phage peptides isolated for PDZ-2 contained in the majority Val and Leu, but also Ile, at the C-terminal position “0.” In contrast to this, none of the PDZ-1 isolates had Val at this position, suggesting that this residue is not preferred at the C terminus. Instead, the majority of these peptides contained Leu, and only a few displayed Ile at this position. In both cases, the vast majority of peptides contained Trp at position –1, and in only a few cases other residues, which were also of a hydrophobic/aromatic nature (Phe, Ser, Ala, Tyr, His, Pro). In contrast, hydrophilic residues were not detected at this –1 position. At additional upstream positions, we did not observe any preferences for specific amino acids.
To determine the molecular recognition preferences of both PDZs, we measured their relative affinities for all isolated artificial peptides using a phage ELISA assay (Fig. 1A). Examination of binding histograms revealed significant differences in recognition profiles between PDZ-1 and PDZ-2 domains. In confirmation of the screening results, peptides containing Val at position 0 did not interact with PDZ-1. However, PDZ-2 interacted with peptides selected by both domains, with visibly higher affinity for those containing C-terminal Val. This indicates higher promiscuity of PDZ-2 than PDZ-1 in the process of ligand recognition.
In addition to this analysis, we performed phage ELISAs for both PDZs using 93 class I artificial peptides containing C-terminal Val, which were isolated for PDZ domain interactors of the synapse-associated protein (SAP) family in our previous work [Kurakin et al., 2007]. We did not detect any interactions between these SAP-PDZ interactors and the Mint1 PDZ-1, which is compatible with the profile described above. Interestingly, the Mint1 PDZ-2 interacted with only 26 of the 93 SAP-PDZ-interactive peptides. The majority of these with high-affinity binding displayed Trp at position –1, similar to what had been observed for peptides interacting with Mint1 PDZs. However, in some cases, residues of polar/hydrophilic or charged character were also present at this position (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material), which indicates that, although preferred, Trp or other hydrophobic residues are not required at the –1 position. Additionally, the fact that only 26 of 93 SAP peptides were positive for binding to Mint1-PDZ-2 clearly shows that, in some cases, the last three positions in the consensus peptide sequence T/S-X-V are not sufficient to determine an interaction, and that other residues located upstream of the C terminus must play an important role in this process, as suggested by other investigators [Niethammer et al., 1998; Birrane et al., 2003; Appleton et al., 2006; Kurakin et al., 2007].
Alignment of the sequences of the various interacting artificial peptides allowed us to deduce a strict consensus sequence, X-W-V/L (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material), which was displayed by the vast majority of the interactors. However, applying less stringent criteria, this consensus could be relaxed and presented as X-W/F-L/I for PDZ-1 and X-Φ-V/L/I for PDZ-2. Furthermore, taking into account the fact that, in some cases, PDZ-2 interacted with peptides containing hydrophilic and polar amino acids at the –1 position, an even more inclusive consensus for the Mint1 PDZ-2 interactors is X-X-V/L/I. A graphical representation of an amino acid multiple sequence alignment (consensus logo) for each PDZ domain, which provides a richer and more precise description than a single consensus sequence identification, is shown in Figure 1B.
Overall, TAIS of the two Mint1 PDZ domains resulted in >1000 positive clones for each domain. Of these, we isolated, sequenced, and tested a set of 88 artificial peptides, and 68 of these 88 displayed the strict consensus of X-W-V/L. This consensus does not fit squarely into any of the three known classes of PDZ interactors (see Discussion), suggesting that the Mint1 PDZ domain interactors represent a variation on the previously described PDZ domain interactors.
The importance of Trp at the –1 position
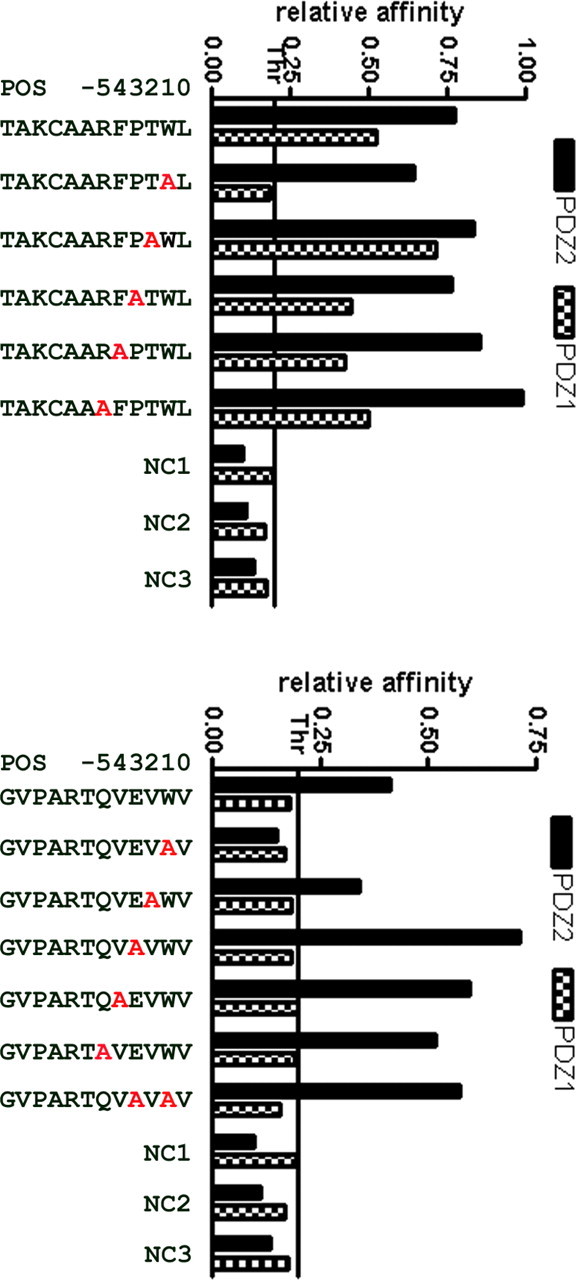
Figure 2 Since Trp was the most highly represented amino acid at the –1 position among all artificial peptides interacting with both PDZ domains, we performed mutagenesis studies to ascertain the necessity, or lack of necessity, for Trp at this position. We chose two peptides, one of which showed relatively high affinity for both PDZ domains (TAKCAARFPTWL) and the other of which showed high affinity for PDZ-2 only (GVPARTQVEVWV). We then synthesized sets of mutant peptides based on these two high-affinity PDZ interactors, in which amino acids at positions from –1 to –5 were replaced by Ala (Figure 2). Relative binding affinities of these peptides for both PDZs were then assessed using ELISA assays. Replacement of Trp by Ala in the TAKCAARFPTWL peptide completely abolished binding to PDZ-1, but did not affect binding to PDZ-2. It is noteworthy that a similar requirement for Trp at the –1 position was reported for the N-type Ca2+ channel (E/D-X-W-C/S) [Maximov et al., 1999]. Ala substitutions at the remaining positions of the TAKCAARFPTWL peptide, however, had smaller effects than the marked effect seen at the –2 position with PDZ-1.
Interaction of PDZ-1 with the GVPARTQVEVWV peptide was not observed. Substitution of Trp at –1 resulted in complete abolition of interaction with PDZ-2. Interestingly, however, an additional, second substitution of Ala for Glu at position –3, resulting in GVPARTQVAVAV, restored and even enhanced binding affinity. Additionally, substitutions at the remaining positions had pronounced positive effects for interaction with PDZ-2. From these experiments, we concluded that Trp plays an important role at position –1 in the test peptides, and we expect that this would be a similar case for the majority of the peptides isolated in our screen. However, we also confirmed that it is not indispensable, as in some cases, its loss could be compensated by substitutions at other positions within a peptide. Our observations support a fine-tuning mechanism of PDZ–peptide interaction by residues other than the last three C-terminal ones, as suggested previously [Kurakin and Bredesen, 2002, 2007; Kurakin et al., 2003].
Identification of novel interactors for Mint1
To identify novel interaction partners for both PDZ domains of the APP-interacting protein Mint1, we first identified candidates by querying the SWISS-PROT and TrEMBL databases using the X-W-V/L consensus, and identified 66 candidates matching this sequence. Additionally, we queried the databases using X-Φ-V/L/I consensus, and from the large set of candidates we randomly selected 20. We used this set of candidates to synthesize a set of 86 dodecamers, which were N-terminally biotinylated proteomic fragments corresponding to the C termini of the predicted natural ligands (supplemental Table 1, available at www.jneurosci.org as supplemental material). These 86 biotinylated 12-mers were then used to confirm or refute the predicted interactions and to evaluate the relative affinities of these natural peptides for each of the two PDZ domains. ELISA assays with GST-PDZ fusions were used as targets.
As positive controls, we synthesized two high-affinity artificial peptides (ASYCKQRNIVWL and GHLSESSQVRWL) and the C-terminal fragment of CCS (Copper chaperone for superoxide dismutase, GRKESAQPPAHL), which has been shown to interact with the Mint1 PDZ-2 [Duquesne et al., 2005]. The C-terminal fragment of the GP41 glycoprotein (CWFSITNWLWYI) was used as a negative control [Maximov et al., 1999]. The binding histograms clearly indicate that PDZ-1 did not bind to peptides containing Val at position 0, just as had been observed in the case of the artificial peptides, whereas PDZ-2 showed higher promiscuity, binding to both peptides containing Val and those containing Leu at the C terminus (Fig. 3A). Interestingly, we did not observe any interaction of PDZ-1 with the CCS-positive control, which has not been reported previously.
To control for nonspecific binding, we performed ELISA assays of all 86 natural peptides using only GST protein as a target. None of the peptides demonstrated an interaction (data not shown). We then identified interactions of 23 of the natural peptides with PDZ-1 and 35 with PDZ-2. Since 12 peptides interacted with both domains, the total number of verified novel interactions was 46. All verified natural ligands are listed in supplemental Table 2 (available at www.jneurosci.org as supplemental material). These interaction partners link Mint1 directly, and APP indirectly, with various cellular functionalities including transport, transcription, metal homeostasis, and molecular motors (Fig. 3B).
Interestingly, 10 of the 46 identified interactors (22%) turned out to be transcriptional regulators (Fig. 3B). In addition to their association with APP, Mint1/X11 family proteins have been implicated in synaptic function, ion channel regulation, and transport, but there has been little discussion of their associations with transcriptional regulators. One study claimed that Mint2 interacted with NF-κB/p65, but a triple protein complex of APP, Mint2, and NF-κB/p65 was not found [Tomita et al., 2000]. Because of the high percentage of transcriptional regulators clustered in our TAIS results, we first focused on two of the transcriptional coactivators: TAZ and YAP.
Both TAZ and YAP interact with Mint1 and Mint3
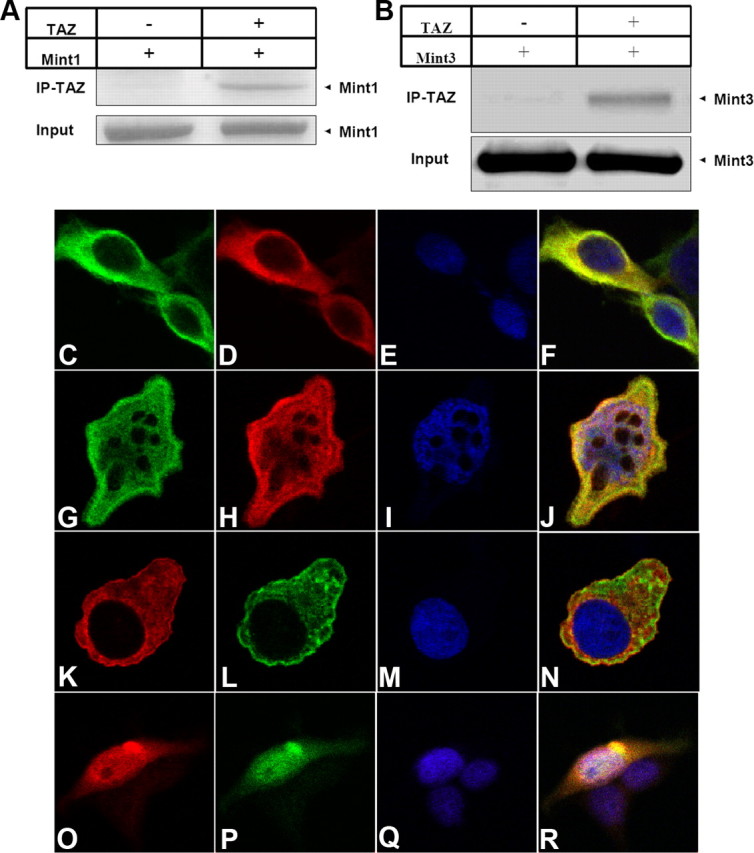
Figure 4 To confirm the interactions between full-length TAZ and Mint1, we cotransfected full-length TAZ with a FLAG tag and full-length Mint1 into 293T cells, and used anti-FLAG antibody to immunoprecipitate TAZ. We found that full-length TAZ did indeed coimmunoprecipitate with Mint1 (Fig. 4A). Since all three members of the Mint1/X11 family display a high level of homology in their PDZ domains, we then asked whether TAZ also interacts with Mint3. We did the same coimmunoprecipitation experiment and found that TAZ did interact with Mint3 (Figure 4B).
It has been shown that Mint1 and Mint2 are distributed mostly in the cytoplasm, whereas Mint3 shuttles between the nucleus and cytoplasm [Sumioka et al., 2008]. Therefore, we asked whether Mint1 might trap TAZ in the cytoplasm, whereas Mint3 allows TAZ to translocate to the nucleus. To address this question, we used immunocytochemistry and confocal microscopy, and we found that, when coexpressed with Mint3, TAZ colocalized with Mint3 and distributed in both cytoplasm and nucleus (Fig. 4G–J). In contrast, when coexpressed with Mint1, TAZ colocalized with Mint1 and was excluded from the nucleus (Fig. 4C–F).
We then performed similar experiments with YAP and obtained similar results. When coexpressed with Mint3, YAP colocalized with Mint3 and distributed in both cytoplasm and nucleus (Fig. 4O–R). However, in the presence of Mint1, YAP colocalized with Mint1 and was trapped in the cytoplasm (Fig. 4K–N).
Transcriptionally active triple protein complexes: APP–Mint3–TAZ and APP–Mint3–YAP
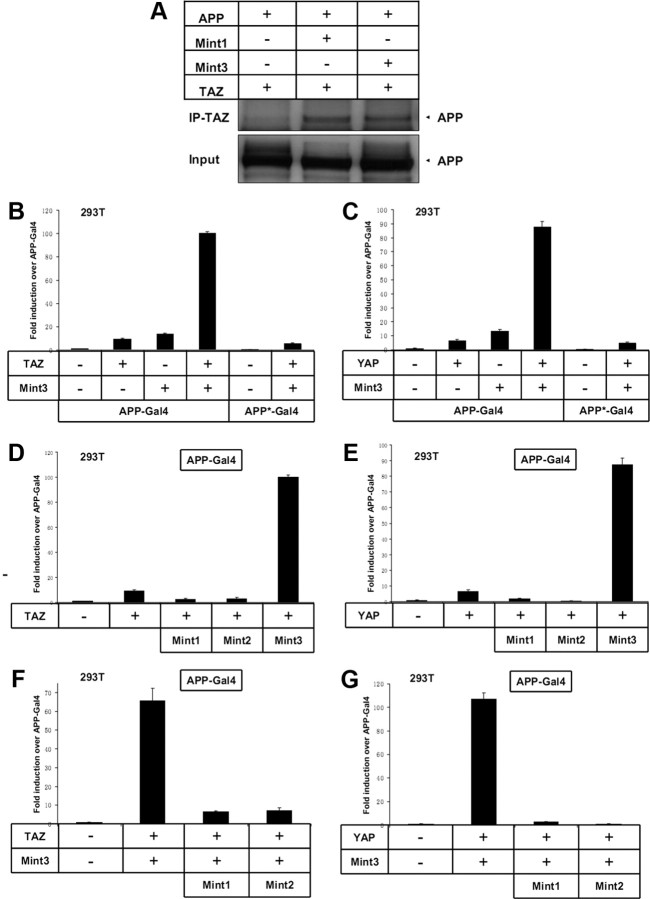
Figure 5 Since Mint1 is a well established binding partner of APP, we asked whether TAZ could associate with APP through Mint1. We coexpressed APP, Mint1, and TAZ (FLAG tagged), and used anti-FLAG antibody to immunoprecipitate TAZ. TAZ coimmunoprecipitated with APP in the presence of Mint1 (Figure 5A), and APP did not coimmunoprecipitate with TAZ in the absence of Mint1. Since TAZ also interacts with Mint3, we then asked whether TAZ could associate with APP through Mint3. Indeed, TAZ coimmunoprecipitated with APP in the presence of Mint3, as well (Figure 5A). Therefore, APP, Mint1/Mint3, and TAZ/YAP form triple protein complexes in transiently cotransfected 293T cells.
To explore the functional link between APP and TAZ, we used a transactivation assay in which the DNA binding domain of Gal4 was fused to APP and transactivation was monitored with a Gal4-dependent reporter plasmid [Cao and Südhof, 2001]. We transfected APP-Gal4 constructs into 293T and B103 cells, and measured transactivation of transcription from a cotransfected Gal4-dependent reporter plasmid expressing luciferase. APP-Gal4 transfected alone was used as a negative control, and all cells were cotransfected with a constitutive β-galactosidase expression plasmid to standardize the transfection efficiency. When we coexpressed TAZ, Mint3, and APP-Gal4, potent transactivation was observed. This transactivation could not be achieved by coexpressing APP-Gal4 with Mint3 only or TAZ only (Fig. 5B). These results suggest that APP, Mint3, and TAZ form a transcriptionally active triple protein complex.
In the cytoplasmic tail of APP, Mint3 binds to the YENPTY motif. To determine whether the binding of Mint3 to APP via the YENPTY motif mediates transactivation, we used an APP-Gal4* construct in which YENPTY had been mutated to YENATA. The mutant APP-Gal4* construct abolishes the interaction between APP and Mint3. The same mutation also abolished the Mint3-dependent stimulation of transcription (Fig. 5B).
To determine whether Mint1 and Mint2 direct similar APP-Gal4 transactivation in the presence of TAZ, we coexpressed TAZ and APP-Gal4 in the presence of Mint1 or Mint2. Neither Mint1 nor Mint2 was able to induce transactivation, indicating that these Mint1/X11 family members play different roles than Mint3 in APP signaling (Fig. 5D).
To define the roles of Mint1 and Mint2 in APP signaling further, we coexpressed Mint1 or Mint2 with APP-Gal4, Mint3, and TAZ. In the presence of Mint1 or Mint2, the transactivation directed by APP-Gal4, Mint3, and TAZ was abolished (Fig. 5F). Thus, Mint3 mediates, whereas Mint1 inhibits, the transactivation of the APP–Mint–TAZ complex.
To determine whether there exists a functional link between YAP and APP, we took a similar approach to that used to evaluate TAZ. When we coexpressed YAP, Mint3, and APP-Gal4, potent transactivation was observed. The potent transactivation could not be achieved by coexpressing APP-Gal4 with YAP alone. When we substituted the APP-Gal4* mutant construct, the potent transactivation directed by YAP was abolished (Fig. 5C). These results indicate that APP, Mint3, and YAP also form a transcriptionally active triple protein complex. We then coexpressed YAP and APP-Gal4 in the presence of Mint1 or Mint2. Mint1 and Mint2 both failed to induce transactivation (Fig. 5E). Similarly, in the presence of Mint1 or Mint2, the transactivation directed by APP-Gal4, Mint3, and YAP was abolished (Fig. 5G).
APP processing regulates the function of TAZ and YAP
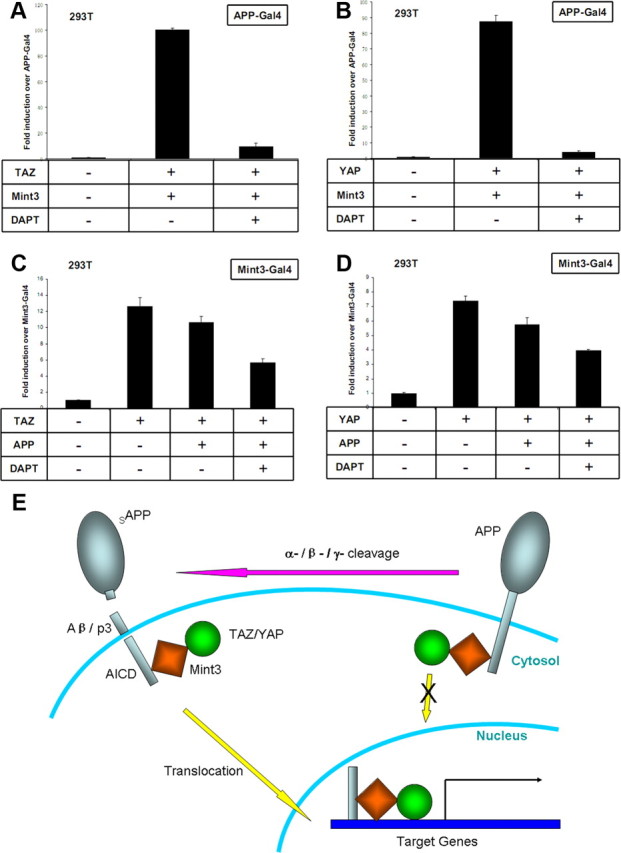
Figure 6 The APP intracellular domain (AICD) is derived by γ-secretase cleavage of APP [Müller et al., 2008]. Since full-length APP is a membrane protein, Mint3 and TAZ attached to APP would be associated with the membrane and excluded from the nucleus. Therefore, regulation of APP γ-secretase cleavage could control the release of the AICD–Mint3–TAZ complex into the nucleus. To test the effect of γ-secretase modulation on TAZ-Mint3-induced APP-Gal4 transactivation, we applied N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), a γ-secretase inhibitor, in the transactivation experiments and found that DAPT treatment abolished the transactivation (Fig. 6A). We did a similar experiment with YAP-Mint3-induced APP-Gal4 transactivation, and DAPT showed the same inhibitory effect (Figure 6B).
A disadvantage of the APP-Gal4 transactivation system is that it only reports the transactivation effects of molecules that associate with APP/AICD, whereas the role that APP itself plays in the signaling events is not clear. To address this problem, we used a Mint3-Gal4 transactivation system in which the DNA binding domain of Gal4 was fused to Mint3 and transactivation was monitored with a Gal4-dependent reporter plasmid. As we expected, coexpression of TAZ with Mint3-Gal4 induced potent transactivation over Mint3-Gal4 alone (Fig. 6C). However, TAZ-induced transactivation was reduced by the coexpression of APP. With DAPT treatment, TAZ induced Mint3-Gal4 transactivation was further decreased in the presence of APP (Fig. 6C). These results indicate that full-length APP trapped Mint3 and TAZ in the cytoplasm, and thereby inhibited the activity of TAZ as a transcriptional coactivator.
To examine the role of APP in YAP induced transactivation, we took a similar approach and again found that YAP induced potent Mint3-Gal4 transactivation. This transactivation was inhibited by APP coexpression and further reduced by DAPT treatment (Fig. 6D).
Discussion
Recent work from multiple laboratories has implicated APP-mediated signaling in the pathophysiology of Alzheimer's disease, but the critical signaling network has not been defined. Here, we used TAIS, a rapid, phage display-based method for the identification of protein–protein interactions and refinement of consensus interaction sequences; this approach resulted in the identification of 46 novel PDZ-interacting proteins that potentially associate with APP through Mint1/X11 family proteins. The profile of potential interactors for the PDZ domains of Mint1 included proteins related to transport, transcription, metal homeostasis, and molecular motors, compatible with previous reports for functional interactors with APP itself. We then confirmed that two transcriptional coactivators, TAZ and YAP, functionally associate with APP through Mint3, and form transcriptionally active triple protein complexes. Furthermore, we found that intact APP localizes Mint3-TAZ and Mint3-YAP to the membrane, excluding them from the nucleus, whereas APP processing releases the AICD–Mint3–TAZ/YAP complexes and allows their translocation into the nucleus, resulting in the activation of target gene transcription.
It should be noted that our observation that the Mint1 PDZ-1 did not select peptides with a C-terminal Val is contrary to a single report indicating such an interaction [Bezprozvanny and Maximov, 2001], in that case with the C-terminal peptide of Neurexin1α (KDEYYV). Two years previously, however, the same authors published work in which they clearly showed a lack of interaction between the Mint1 PDZ-1 and Neurexin1α C-peptide [Maximov et al., 1999]. Therefore, additional work is needed to determine whether this case represents an exception to the general pattern disclosed by our studies that the Mint1 PDZ-1 does not associate with peptides or proteins that display a C-terminal Val. In contrast to this pattern for the Mint1 PDZ-1, however, our data suggest that the Mint1 PDZ-2 should interact with the C-peptide of Neurexin.
With the interactions of a growing number of PDZ domains being characterized, it is becoming clear that at least some PDZ domains interact with multiple classes of peptide ligands. A dual specificity of the Mint1 PDZ-1 has been suggested by Bezprozvanny and Maximov [2001], since they demonstrated its interaction with the C-terminal peptides of N-type Ca2+ channels (E/D-X-W-C/S), which are defined as a novel class III [Maximov et al., 1999], as well as with the C-terminal peptide of Neurexin1α (KDEYYV), which is assigned to class II [Bezprozvanny and Maximov, 2001]. To date, there have been only a few natural ligands identified for the PDZ domains of Mint1 [Rogelj et al., 2006]. Although their sequences may provide some clues to the Mint1 PDZ specificity and classification, a greater number of ligands need to be identified and analyzed to provide more accurate characterization of these domains. Analysis of 88 artificial peptides isolated in our screen revealed the presence of sequences assigned to all three classes of PDZ domain interactors: class I (T/S-X-Φ, where Φ is a hydrophobic residue), class II (Φ-X-Φ), and class III (D/E-X-V) [Vaccaro and Dente, 2002].
However, in addition to peptides from these three classes, we also identified peptides containing positively charged amino acids (R, K), as well as polar but uncharged or weakly charged amino acids (N, Q, H) at position –2. These peptides do not match any of the PDZ classes defined so far, suggesting an extension of the classification. These unclassified peptides are closer to fitting into class III than into class I or II, and if the criteria for class III were relaxed to include polar/hydrophilic residues at position –2 (shown as class III+ in supplemental Fig. 1A, available at www.jneurosci.org as supplemental material), then these peptides would meet such relaxed criteria for an expanded class III. However, if the current criteria were applied strictly, then these peptides would represent a novel class of PDZ-interacting peptides.
The TAIS approach allowed an accurate prediction of novel consensus interaction sequences for the PDZ domains of Mint1 and thus a prediction of natural ligands for these domains. Synthesis of peptides representing the C termini of the predicted partners resulted in the confirmation of 46 novel interactors, 10 of which are transcriptional coactivators. These novel interactors represent new candidate mediators of the Alzheimer-related signaling abnormality. Among these, two were chosen for additional studies: TAZ because of its relationship to the PPAR-γ signaling pathway implicated in Alzheimer's disease, and YAP because of its role in setting organ size (see below). By analogy to the tripartite complex of APP–Fe65–Tip60 [Müller et al., 2008], in the current study we identified two novel transcriptionally active triple protein complexes: APP–Mint3–TAZ and APP–Mint3–YAP. However, the current findings show clear distinctions from those of the APP–Fe65–Tip60 complex: first, Fe65 is capable of inducing transactivation in the absence of Tip60, and one study showed that Tip60 inhibited Fe65-induced transactivation [Yang et al., 2006]. In contrast, Mint3 did not display transactivation unless TAZ or YAP was also present. Second, the presence of APP is required for Fe65 to induce Tip60-Gal4 transactivation. In contrast, intact APP actually inhibited the activities of Mint3-TAZ and Mint3-YAP. Third, Fe65 is expressed predominantly in the brain [Simeone et al., 1994], whereas both APP and Mint3 are expressed ubiquitously (which is compatible with a general physiological function of APP-Mint3 signaling). Mint1 and Mint2, however, are expressed mostly in neurons, and thus appear to be neural-specific inhibitory modulators of the APP-Mint3 signaling.
The lack of transactivation with Mint1 and Mint2—and their inhibition of the Mint3–YAP/TAZ–APP transactivation—is compatible with the lack of nuclear translocation in the presence of Mint1 and Mint2, which are both located predominantly in the cytoplasm. This supports previous studies showing that Mint1 and Mint2, but not Mint3, strongly inhibit transactivation directed by APP–Gal4–VP16 [Biederer et al., 2002].
TAZ and YAP are transcriptional coactivators that share high levels of sequence similarity with each other. Both TAZ and YAP can be sequestered in the cytoplasm and rendered functionally inactive by binding to 14-3-3 proteins in a phosphorylation-dependent manner [Hong and Yaffe, 2006]. Here, we have shown that Mint1 dominantly controls the location of TAZ and YAP by excluding them from the nucleus. In contrast, although Mint3 shuttles between the cytoplasm and nucleus, coexpression of APP inhibited the transactivation directed by Mint3-TAZ/YAP, and the transactivation was further decreased by abolishing the γ-secretase cleavage of APP. These results fit well with a model in which intact APP traps Mint3 and TAZ/YAP in the cytoplasm, whereas γ-secretase cleavage releases the triple protein complexes of AICD–Mint3–TAZ/YAP and allows them to translocate into the nucleus and induce transcription of target genes (Fig. 6E).
To date, there has not been any report linking TAZ or YAP to AD, but there are some intriguing clues to their potential involvement: TAZ has been negatively implicated in PPARγ signaling activation [Hong et al., 2005] and positively implicated in TGFβ/Smad signaling [Varelas et al., 2008]. Since PPARγ signaling could block AD pathogenesis by inhibiting neuroinflammation [Kummer and Heneka, 2008], whereas TGFβ/Smad signaling could promote AD pathogenesis by enhancing neuroinflammation [Town et al., 2008], it is possible that TAZ serves as a link between Aβ activated APP signaling and downstream gene activation involved in AD progression. YAP, however, has been identified as the key mediator of organ size control [Dong et al., 2007]. One major feature of AD is progressive brain atrophy, with dentate gyrus atrophy at a very early stage—in an AD mouse model, this atrophy precedes synapse and neuronal loss, leaving the mechanism of this early atrophy unexplained [Galvan et al., 2006]. This raises an intriguing possibility: that the early atrophy in Alzheimer's disease may actually be a result of signaling rather than a secondary event, and YAP may serve as a key mediator of early brain atrophy directed through the Aβ-APP signaling pathway.
Although TAZ and YAP share a high level of structure similarity, their functions are quite different. The C-terminal transcriptional regulatory domain of both TAZ and YAP, in isolation, can strongly coactivate gene expression when corecruited with Runx1 to an artificial promoter in a GAL4-driven luciferase reporter system. In vivo, however, TAZ and YAP appear, instead, to regulate Runx2-driven genes during osteoblast differentiation in opposing ways [Kanai et al., 2000; Zaidi et al., 2004; Hong et al., 2005]. Therefore, it is possible that TAZ and YAP act antagonistically in AD pathogenesis (TAZ being pro-AD and YAP anti-AD), even though they both induce potent APP-Gal4 transactivation. Thus, additional study is required to elucidate possible roles that YAP and TAZ might play in the pathogenesis of AD, and ongoing efforts are being made to address this question.
Acknowledgments
This work was supported by a Douglas and Ellen Rosenberg Foundation Fellowship (Q.Z.), by The Keck Foundation, and by National Institutes of Health Grant NS45093 (D.E.B.). A.K. is supported by National Institute on Aging Grant AG031380. We thank Drs. Patrick Mehlen, Veronique Corset, Thomas Südhof, Marius Sudol, Michael Yaffe, Iain Farrance, and Toshiharu Suzuki for providing plasmids.
References:
Appleton BA, Zhang Y, Wu P, Yin JP, Hunziker W, Skelton NJ, Sidhu SS, Wiesmann C.
Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1.
Insights into determinants of PDZ domain specificity.
J Biol Chem. 2006;281:22312–22320Ashley J, Packard M, Ataman B, Budnik V.
Fasciclin II signals new synapse formation through amyloid precursor protein and the scaffolding protein dX11/Mint.
J Neurosci. 2005;25:5943–5955Barnham KJ, Bush AI.
Metals in Alzheimer's and Parkinson's diseases.
Curr Opin Chem Biol. 2008;12:222–228Bezprozvanny I, Maximov A.
Classification of PDZ domains.
FEBS Lett. 2001;509:457–462Biederer T, Cao X, Südhof TC, Liu X.
Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms.
J Neurosci. 2002;22:7340–7351Birrane G, Chung J, Ladias JA.
Novel mode of ligand recognition by the Erbin PDZ domain.
J Biol Chem. 2003;278:1399–1402Butterfield DA.
Amyloid beta-peptide [1-42]-associated free radical-induced oxidative stress and neurodegeneration
in Alzheimer's disease brain: mechanisms and consequences.
Curr Med Chem. 2003;10:2651–2659Butz S, Okamoto M, Südhof TC.
A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain.
Cell. 1998;94:773–782Cao X, Südhof TC.
A transcriptionally [correction of transcriptively] active complex of APP with Fe65
and histone acetyltransferase Tip60.
Science. 2001;293:115–120Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA.
Elucidation of a universal size-control mechanism in Drosophila and mammals.
Cell. 2007;130:1120–1133Duquesne AE, Ruijter M, Brouwer J, Drijfhout JW, Nabuurs SB, Spronk CA.
Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with
the C-terminal peptide of the human copper chaperone for superoxide dismutase.
J Biomol NMR. 2005;32:209–218Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Bredesen DE.
Reversal of Alzheimer's-like Pathology and Behavior in Human APP Transgenic Mice
by Mutation of Asp664
Proc Natl Acad Sci U S A. 2006 (May 2); 103 (18): 7130–7135Glabe C.
Intracellular mechanisms of amyloid accumulation and pathogenesis in Alzheimer's disease.
J Mol Neurosci. 2001;17:137–145Ho A, Liu X, Südhof TC.
Deletion of Mint proteins decreases amyloid production in transgenic mouse models of Alzheimer's disease.
J Neurosci. 2008;28:14392–14400Hong JH, Yaffe MB.
TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation.
Cell Cycle. 2006;5:176–179Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E.
TAZ, a transcriptional modulator of mesenchymal stem cell differentiation.
Science. 2005;309:1074–1078Hsia AY, Masliah E, McConlogue L, Yu GQ, Tatsuno G, Hu K, Kholodenko D, Malenka RC.
Plaque-independent disruption of neural circuits in Alzheimer's disease mouse models.
Proc Natl Acad Sci U S A. 1999;96:3228–3233Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A.
TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins.
EMBO J. 2000;19:6778–6791King GD, Turner RS.
Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer's disease risk?
Exp Neurol. 2004;185:208–219King GD, Cherian K, Turner RS.
X11alpha impairs gamma- but not beta-cleavage of amyloid precursor protein.
J Neurochem. 2004;88:971–982Kummer MP, Heneka MT.
PPARs in Alzheimer's disease.
PPAR Res. 2008;2008:403896Kurakin A, Bredesen D.
Target-assisted iterative screening reveals novel interactors for PSD95, Nedd4, Src, Abl and Crk proteins.
J Biomol Struct Dyn. 2002;19:1015–1029Kurakin A, Bredesen DE.
An unconventional IAP-binding motif revealed by target-assisted iterative screening (TAIS)
of the BIR3-cIAP1 domain.
J Mol Recognit. 2007;20:39–50Kurakin A, Wu S, Bredesen DE.
Target-assisted iterative screening of phage surface display cDNA libraries.
Methods Mol Biol. 2004;264:47–60Kurakin A, Swistowski A, Wu SC, Bredesen DE.
The PDZ domain as a complex adaptive system.
PLoS One. 2007;2:e953Kurakin AV, Wu S, Bredesen DE.
Atypical recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by target-assisted iterative screening.
J Biol Chem. 2003;278:34102–34109Lee JH, Lau KF, Perkinton MS, Standen CL, Shemilt SJ, Mercken L, Cooper JD.
The neuronal adaptor protein X11alpha reduces Abeta levels in the brains of Alzheimer's APPswe Tg2576 transgenic mice.
J Biol Chem. 2003;278:47025–47029Lee JH, Lau KF, Perkinton MS, Standen CL, Rogelj B, Falinska A, McLoughlin DM.
The neuronal adaptor protein X11beta reduces amyloid beta-protein levels and amyloid plaque
formation in the brains of transgenic mice.
J Biol Chem. 2004;279:49099–49104Lourenco FC, Galvan V, Fombonne J, Corset V, Llambi F, Muller U, Bredesen DE, Mehlen P.
Netrin-1 Interacts with Amyloid Precursor Protein and Regulates Amyloid-beta Production
Cell Death Differ. 2009 (May); 16 (5): 655–663Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE.
A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor.
Nat Med. 2000;6:397–404Ma QL, Yang F, Calon F, Ubeda OJ, Hansen JE, Weisbart RH, Beech W, Frautschy SA, Cole GM.
p21-activated kinase-aberrant activation and translocation in Alzheimer disease pathogenesis.
J Biol Chem. 2008;283:14132–14143Maximov A, Südhof TC, Bezprozvanny I.
Association of neuronal calcium channels with modular adaptor proteins.
J Biol Chem. 1999;274:24453–24456Mueller HT, Borg JP, Margolis B, Turner RS.
Modulation of amyloid precursor protein metabolism by X11alpha/Mint-1. A deletion analysis of
protein-protein interaction domains.
J Biol Chem. 2000;275:39302–39306Müller T, Meyer HE, Egensperger R, Marcus K.
The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression,
apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease.
Prog Neurobiol. 2008;85:393–406Nguyen TV, Galvan V, Huang W, Banwait S, Tang H, Zhang J, Bredesen DE.
Signal transduction in Alzheimer disease: p21-activated kinase signaling requires C-terminal cleavage
of APP at Asp664.
J Neurochem. 2008;104:1065–1080Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg RJ, Craig AM, Sheng M.
CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90.
Neuron. 1998;20:693–707Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M.
APP binds DR6 to trigger axon pruning and neuron death via distinct caspases.
Nature. 2009;457:981–989Rogelj B, Mitchell JC, Miller CC, McLoughlin DM.
The X11/Mint family of adaptor proteins.
Brain Res Rev. 2006;52:305–315Saito Y, Sano Y, Vassar R, Gandy S, Nakaya T, Yamamoto T, Suzuki T.
X11 proteins regulate the translocation of amyloid beta-protein precursor (APP) into detergent-resistant
membrane and suppress the amyloidogenic cleavage of APP by beta-site-cleaving enzyme in brain.
J Biol Chem. 2008;283:35763–35771Setou M, Nakagawa T, Seog DH, Hirokawa N.
Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport.
Science. 2000;288:1796–1802Simeone A, Duilio A, Fiore F, Acampora D, De Felice C, Faraonio R, Paolocci F.
Expression of the neuron-specific FE65 gene marks the development of embryo ganglionic derivatives.
Dev Neurosci. 1994;16:53–60Sumioka A, Saito Y, Sakuma M, Araki Y, Yamamoto T, Suzuki T.
The X11L/X11beta/MINT2 and X11L2/X11gamma/MINT3 scaffold proteins shuttle between the nucleus and cytoplasm.
Exp Cell Res. 2008;314:1155–1162Tomita S, Fujita T, Kirino Y, Suzuki T.
PDZ domain-dependent suppression of NF-kappaB/p65-induced Abeta42 production by a neuron-specific X11-like protein.
J Biol Chem. 2000;275:13056–13060Town T, Laouar Y, Pittenger C, Mori T, Szekely CA, Tan J, Duman RS, Flavell RA.
Blocking TGF-beta-Smad2/3 innate immune signaling mitigates Alzheimer-like pathology.
Nat Med. 2008;14:681–687Townsend M, Mehta T, Selkoe DJ.
Soluble Abeta inhibits specific signal transduction cascades common to the insulin receptor pathway.
J Biol Chem. 2007;282:33305–33312Vaccaro P, Dente L.
PDZ domains: troubles in classification.
FEBS Lett. 2002;512:345–349Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J.
TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal.
Nat Cell Biol. 2008;10:837–848Yang Z, Cool BH, Martin GM, Hu Q.
A dominant role for FE65 (APBB1) in nuclear signaling.
J Biol Chem. 2006;281:4207–4214Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS.
Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription.
EMBO J. 2004;23:790–799Zhao L, Ma QL, Calon F, Harris-White ME, Yang F, Lim GP, Morihara T, Ubeda OJ.
Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease.
Nat Neurosci. 2006;9:234–242.
Return to ALZHEIMER's
Since 4-10-2020
| Home Page | Visit Our Sponsors | Become a Sponsor |
Please read our DISCLAIMER |